Positive Transfer Of The Whisper Speech Transformer To Human And Animal Voice Activity Detection
We proposed WhisperSeg, utilizing the Whisper Transformer pre-trained for Automatic Speech Recognition (ASR) for both human and animal Voice Activity Detection (VAD). For more details, please refer to our paper
Positive Transfer of the Whisper Speech Transformer to Human and Animal Voice Activity Detection
Nianlong Gu, Kanghwi Lee, Maris Basha, Sumit Kumar Ram, Guanghao You, Richard H. R. Hahnloser <br> University of Zurich and ETH Zurich
The model "nccratliri/whisperseg-large-ms" is the checkpoint of the multi-species WhisperSeg-large that was finetuned on the vocal segmentation datasets of five species.
Usage
Clone the GitHub repo and install dependencies
git clone https://github.com/nianlonggu/WhisperSeg.git
cd WhisperSeg; pip install -r requirements.txt
Then in the folder "WhisperSeg", run the following python script:
from model import WhisperSegmenter
import librosa
import json
segmenter = WhisperSegmenter( "nccratliri/whisperseg-large-ms", device="cuda" )
sr = 32000
min_frequency = 0
spec_time_step = 0.0025
min_segment_length = 0.01
eps = 0.02
num_trials = 3
audio, _ = librosa.load( "data/example_subset/Zebra_finch/test_adults/zebra_finch_g17y2U-f00007.wav",
sr = sr )
prediction = segmenter.segment( audio, sr = sr, min_frequency = min_frequency, spec_time_step = spec_time_step,
min_segment_length = min_segment_length, eps = eps,num_trials = num_trials )
print(prediction)
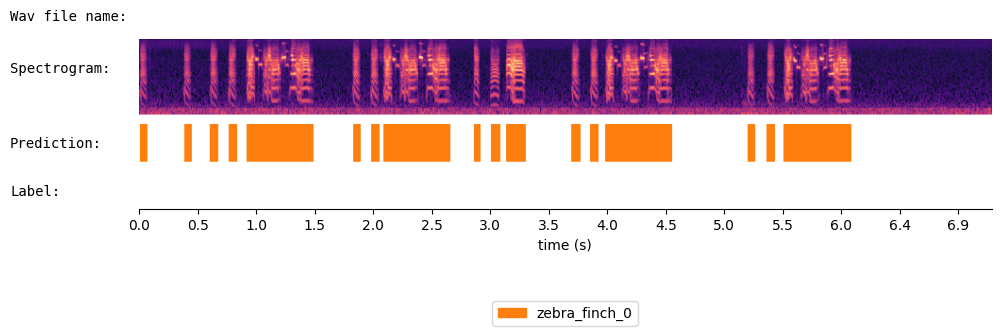
{'onset': [0.01, 0.38, 0.603, 0.758, 0.912, 1.813, 1.967, 2.073, 2.838, 2.982, 3.112, 3.668, 3.828, 3.953, 5.158, 5.323, 5.467], 'offset': [0.073, 0.447, 0.673, 0.83, 1.483, 1.882, 2.037, 2.643, 2.893, 3.063, 3.283, 3.742, 3.898, 4.523, 5.223, 5.393, 6.043], 'cluster': ['zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0', 'zebra_finch_0']}
Visualize the results of WhisperSeg:
from audio_utils import SpecViewer
spec_viewer = SpecViewer()
spec_viewer.visualize( audio = audio, sr = sr, min_frequency= min_frequency, prediction = prediction,
window_size=8, precision_bits=1
)
Run it in Google Colab: <a href="https://colab.research.google.com/github/nianlonggu/WhisperSeg/blob/master/docs/WhisperSeg_Voice_Activity_Detection_Demo.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a> For more details, please refer to the GitHub repository: https://github.com/nianlonggu/WhisperSeg
Citation
When using our code or models for your work, please cite the following paper:
@article {Gu2023.09.30.560270,
author = {Nianlong Gu and Kanghwi Lee and Maris Basha and Sumit Kumar Ram and Guanghao You and Richard Hahnloser},
title = {Positive Transfer of the Whisper Speech Transformer to Human and Animal Voice Activity Detection},
elocation-id = {2023.09.30.560270},
year = {2023},
doi = {10.1101/2023.09.30.560270},
publisher = {Cold Spring Harbor Laboratory},
abstract = {This paper introduces WhisperSeg, utilizing the Whisper Transformer pre-trained for Automatic Speech Recognition (ASR) for human and animal Voice Activity Detection (VAD). Contrary to traditional methods that detect human voice or animal vocalizations from a short audio frame and rely on careful threshold selection, WhisperSeg processes entire spectrograms of long audio and generates plain text representations of onset, offset, and type of voice activity. Processing a longer audio context with a larger network greatly improves detection accuracy from few labeled examples. We further demonstrate a positive transfer of detection performance to new animal species, making our approach viable in the data-scarce multi-species setting.Competing Interest StatementThe authors have declared no competing interest.},
URL = {https://www.biorxiv.org/content/early/2023/10/02/2023.09.30.560270},
eprint = {https://www.biorxiv.org/content/early/2023/10/02/2023.09.30.560270.full.pdf},
journal = {bioRxiv}
}
Contact
nianlong.gu@uzh.ch