This model is a fine-tuned model of BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext (hugging-face card). The current model was developed for the web-based ANDDigest system for the classification of the short names of cell components in texts on the basis of their context (the name considered to be short if it's length is 4 symbols or less). The analyzed name should be replaced in text with <andsystem-candidate> tag.<br> <br> <b>Input:</b><br> Any biomedical text where a name of classified object is replaced with <andsystem-candidate> tag, for example, this pubmed abstract:<br> <i>Merkel cell carcinoma in lymph nodes with and without primary origin. The prognosis of <b><andsystem-candidate></b> with lymph node involvement was better in patients with an unknown than a known primary. Treatment with a uniform aggressive combined chemoradiation regimen, with or without lymphadenectomy, led to better survival rates than previously reported.</i> <br> <br>In this example <i>MCC</i> abbreviation, which refers to the Merkel cell carcinoma, was replaced with <i><andsystem-candidate></i>. Please keep in mind that maximum length of input sequence for BERT is limited to 512 tokens. <br> <b>Output:</b><br> <i>LABEL_0</i> refers to the probability of the <i>FALSE</i> recognition, i.e. if the context of <andsystem-candidate> doesn't corresponds to the context specific for cell components.<br> <i>LABEL_1</i> refers to the probability of the <i>TRUE</i> recognition, i.e. when the context of <andsystem-candidate> corresponds to the context specific for cell components.<br> <br>
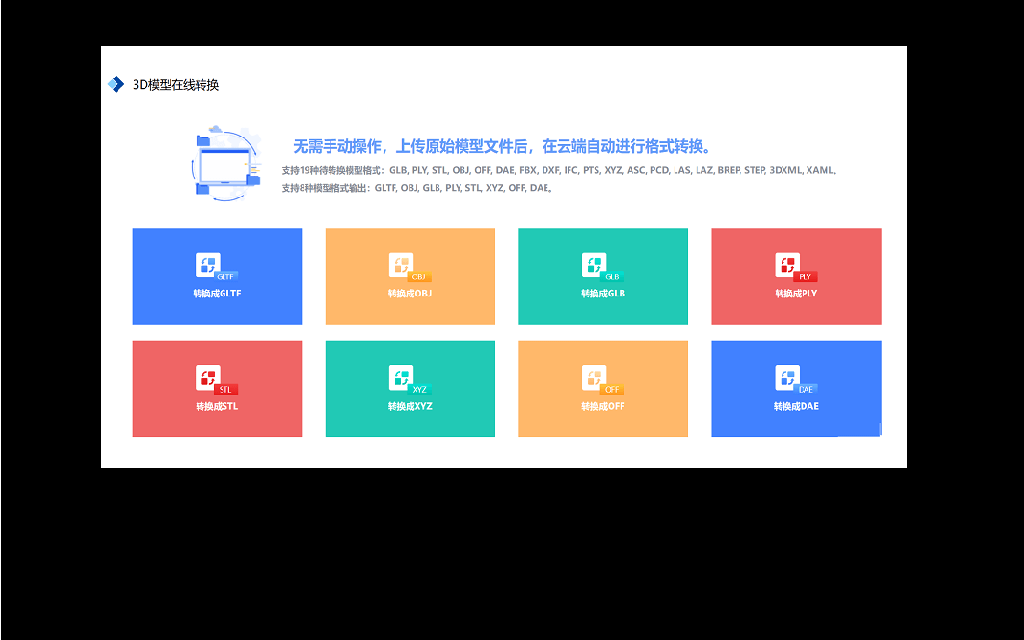
NSDT 3DConvert
Convert 30+ 3D formats online: GLTF, GLB, GBX, OBJ, DAE, IFC, STEP, STL...
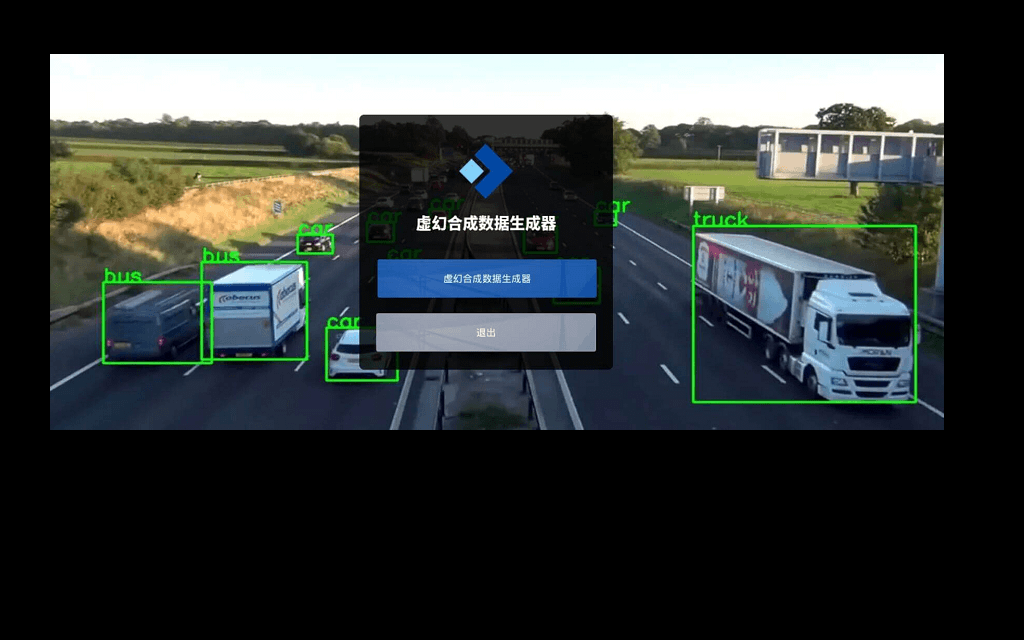
UnrealSynth
Unreal engine based photo realistic synthetic data generator for YOLO.

DreamTexture.js
AI powered 3d texture generation and projection SDK for three.js.